Converting masks back to annotations¶
Overview:

Most segmentation algorithms produce outputs in an image format. Visualizing these outputs in HistomicsUI requires conversion from mask images to an annotation document containing (x,y) coordinates in the whole-slide image coordinate frame. This notebook demonstrates this conversion process in two steps:
Converting a mask image into contours (coordinates in the mask frame)
Placing contours data into a format following the annotation document schema that can be pushed to DSA for visualization in HistomicsUI.
This notebook is based on work described in Amgad et al, 2019:
Mohamed Amgad, Habiba Elfandy, Hagar Hussein, …, Jonathan Beezley, Deepak R Chittajallu, David Manthey, David A Gutman, Lee A D Cooper, Structured crowdsourcing enables convolutional segmentation of histology images, Bioinformatics, 2019, btz083
Where to look?
|_ histomicstk/
|_annotations_and_masks/
| |_masks_to_annotations_handler.py
|_tests/
|_test_masks_to_annotations_handler.py
[1]:
import os
CWD = os.getcwd()
import girder_client
from pandas import read_csv
from imageio import imread
from histomicstk.annotations_and_masks.masks_to_annotations_handler import (
get_contours_from_mask,
get_single_annotation_document_from_contours,
get_annotation_documents_from_contours)
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = 7, 7
1. Connect girder client and set parameters¶
[2]:
# APIURL = 'http://demo.kitware.com/histomicstk/api/v1/'
# SAMPLE_SLIDE_ID = '5bbdee92e629140048d01b5d'
APIURL = 'http://candygram.neurology.emory.edu:8080/api/v1/'
SAMPLE_SLIDE_ID = '5d586d76bd4404c6b1f286ae'
# Connect to girder client
gc = girder_client.GirderClient(apiUrl=APIURL)
gc.authenticate(interactive=True)
# gc.authenticate(apiKey='kri19nTIGOkWH01TbzRqfohaaDWb6kPecRqGmemb')
[2]:
{'_accessLevel': 2,
'_id': '59bc677892ca9a0017c2e855',
'_modelType': 'user',
'admin': True,
'created': '2017-09-15T23:51:20.203000+00:00',
'email': 'mtageld@emory.edu',
'emailVerified': False,
'firstName': 'Mohamed',
'groupInvites': [],
'groups': ['59f7713a92ca9a0017a29765',
'5c607488e62914004d0ff4a6',
'5e44a2e0ddda5f8398785304',
'5e76b3f3ddda5f83982beb9a'],
'lastName': 'Tageldin',
'login': 'kheffah',
'otp': False,
'public': True,
'size': 0,
'status': 'enabled'}
Let’s inspect the ground truth codes file¶
This contains the ground truth codes and information dataframe. This is a dataframe that is indexed by the annotation group name and has the following columns:
group: group name of annotation (string), eg. “mostly_tumor”GT_code: int, desired ground truth code (in the mask) Pixels of this value belong to corresponding group (class)color: str, rgb format. eg. rgb(255,0,0).
NOTE:
Zero pixels have special meaning and do not encode specific ground truth class. Instead, they simply mean ‘Outside ROI’ and should be ignored during model training or evaluation.
[3]:
# read GTCodes dataframe
GTCODE_PATH = os.path.join(
CWD, '..', '..', 'tests', 'test_files', 'sample_GTcodes.csv')
GTCodes_df = read_csv(GTCODE_PATH)
GTCodes_df.index = GTCodes_df.loc[:, 'group']
[4]:
GTCodes_df.head()
[4]:
| group | overlay_order | GT_code | is_roi | is_background_class | color | comments | |
|---|---|---|---|---|---|---|---|
| group | |||||||
| roi | roi | 0 | 254 | 1 | 0 | rgb(200,0,150) | NaN |
| evaluation_roi | evaluation_roi | 0 | 253 | 1 | 0 | rgb(255,0,0) | NaN |
| mostly_tumor | mostly_tumor | 1 | 1 | 0 | 0 | rgb(255,0,0) | core class |
| mostly_stroma | mostly_stroma | 2 | 2 | 0 | 1 | rgb(255,125,0) | core class |
| mostly_lymphocytic_infiltrate | mostly_lymphocytic_infiltrate | 1 | 3 | 0 | 0 | rgb(0,0,255) | core class |
Read and visualize mask¶
[5]:
# read mask
X_OFFSET = 59206
Y_OFFSET = 33505
MASKNAME = 'TCGA-A2-A0YE-01Z-00-DX1.8A2E3094-5755-42BC-969D-7F0A2ECA0F39' + \
'_left-%d_top-%d_mag-BASE.png' % (X_OFFSET, Y_OFFSET)
MASKPATH = os.path.join(CWD, '..', '..', 'tests', 'test_files', 'annotations_and_masks', MASKNAME)
MASK = imread(MASKPATH)
[6]:
plt.figure(figsize=(7,7))
plt.imshow(MASK)
plt.title(MASKNAME[:23])
plt.show()
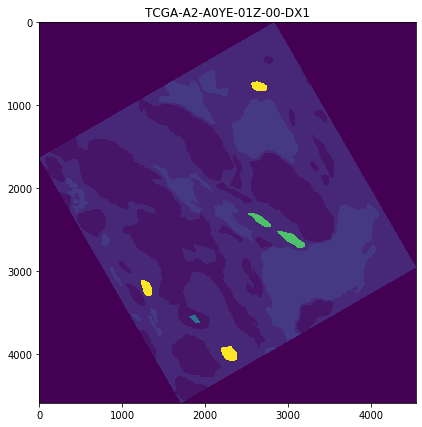
2. Get contours from mask¶
This function get_contours_from_mask() generates contours from a mask image. There are many parameters that can be set but most have defaults set for the most common use cases. The only required parameters you must provide are MASK and GTCodes_df, but you may want to consider setting the following parameters based on your specific needs: get_roi_contour, roi_group, discard_nonenclosed_background, background_group, that control behaviour regarding region of interest
(ROI) boundary and background pixel class (e.g. stroma).
[7]:
print(get_contours_from_mask.__doc__)
Parse ground truth mask and gets contours for annotations.
Parameters
-----------
MASK : nd array
ground truth mask (m,n) where pixel values encode group membership.
GTCodes_df : pandas Dataframe
the ground truth codes and information dataframe.
This is a dataframe that is indexed by the annotation group name and
has the following columns.
group: str
group name of annotation, eg. mostly_tumor.
GT_code: int
desired ground truth code (in the mask). Pixels of this value
belong to corresponding group (class).
color: str
rgb format. eg. rgb(255,0,0).
groups_to_get : None
if None (default) then all groups (ground truth labels) will be
extracted. Otherwise pass a list of strings like ['mostly_tumor',].
MIN_SIZE : int
minimum bounding box size of contour
MAX_SIZE : None
if not None, int. Maximum bounding box size of contour. Sometimes
very large contours cause segmentation faults that originate from
opencv and are not caught by python, causing the python process
to unexpectedly hault. If you would like to set a maximum size to
defend against this, a suggested maximum would be 15000.
get_roi_contour : bool
whether to get contour for boundary of region of interest (ROI). This
is most relevant when dealing with multiple ROIs per slide and with
rotated rectangular or polygonal ROIs.
roi_group : str
name of roi group in the GT_Codes dataframe (eg roi)
discard_nonenclosed_background : bool
If a background group contour is NOT fully enclosed, discard it.
This is a purely aesthetic method, makes sure that the background group
contours (eg stroma) are discarded by default to avoid cluttering the
field when posted to DSA for viewing online. The only exception is
if they are enclosed within something else (eg tumor), in which case
they are kept since they represent holes. This is related to
https://github.com/DigitalSlideArchive/HistomicsTK/issues/675
WARNING - This is a bit slower since the contours will have to be
converted to shapely polygons. It is not noticeable for hundreds of
contours, but you will notice the speed difference if you are parsing
thousands of contours. Default, for this reason, is False.
background_group : str
name of background group in the GT_codes dataframe (eg mostly_stroma)
verbose : bool
Print progress to screen?
monitorPrefix : str
text to prepend to printed statements
Returns
--------
pandas DataFrame
contours extracted from input mask. The following columns are output.
group : str
annotation group (ground truth label).
color : str
annotation color if it were to be posted to DSA.
is_roi : bool
whether this annotation is a region of interest boundary
ymin : int
minimum y coordinate
ymax : int
maximum y coordinate
xmin : int
minimum x coordinate
xmax : int
maximum x coordinate
has_holes : bool
whether this contour has holes
touches_edge-top : bool
whether this contour touches top mask edge
touches_edge-bottom : bool
whether this contour touches bottom mask edge
touches_edge-left : bool
whether this contour touches left mask edge
touches_edge-right : bool
whether this contour touches right mask edge
coords_x : str
vertex x coordinates comma-separated values
coords_y
vertex y coordinated comma-separated values
Extract contours¶
[8]:
# Let's extract all contours from a mask, including ROI boundary. We will
# be discarding any stromal contours that are not fully enclosed within a
# non-stromal contour since we already know that stroma is the background
# group. This is so things look uncluttered when posted to DSA.
groups_to_get = None
contours_df = get_contours_from_mask(
MASK=MASK, GTCodes_df=GTCodes_df, groups_to_get=groups_to_get,
get_roi_contour=True, roi_group='roi',
discard_nonenclosed_background=True,
background_group='mostly_stroma',
MIN_SIZE=30, MAX_SIZE=None, verbose=True,
monitorPrefix=MASKNAME[:12] + ': getting contours')
TCGA-A2-A0YE: getting contours: non-roi: roi: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: evaluation_roi: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: mostly_tumor: getting contours
TCGA-A2-A0YE: getting contours: non-roi: mostly_tumor: adding contours
TCGA-A2-A0YE: getting contours: non-roi: mostly_stroma: getting contours
TCGA-A2-A0YE: getting contours: non-roi: mostly_stroma: adding contours
TCGA-A2-A0YE: getting contours: non-roi: nest 1 of 11: TOO SIMPLE (1 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 2 of 11: TOO SIMPLE (2 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 3 of 11: TOO SIMPLE (1 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 4 of 11: TOO SIMPLE (1 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 5 of 11: TOO SMALL (10 x 18 pixels) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 6 of 11: TOO SIMPLE (1 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 8 of 11: TOO SIMPLE (1 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: nest 9 of 11: TOO SIMPLE (1 coordinates) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: mostly_lymphocytic_infiltrate: getting contours
TCGA-A2-A0YE: getting contours: non-roi: mostly_lymphocytic_infiltrate: adding contours
TCGA-A2-A0YE: getting contours: non-roi: nest 5 of 14: TOO SMALL (23 x 74 pixels) -- IGNORED
TCGA-A2-A0YE: getting contours: non-roi: necrosis_or_debris: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: glandular_secretions: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: mostly_blood: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: exclude: getting contours
TCGA-A2-A0YE: getting contours: non-roi: exclude: adding contours
TCGA-A2-A0YE: getting contours: non-roi: metaplasia_NOS: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: mostly_fat: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: mostly_plasma_cells: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: other_immune_infiltrate: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: mostly_mucoid_material: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: normal_acinus_or_duct: getting contours
TCGA-A2-A0YE: getting contours: non-roi: normal_acinus_or_duct: adding contours
TCGA-A2-A0YE: getting contours: non-roi: lymphatics: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: undetermined: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: nerve: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: skin_adnexia: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: blood_vessel: getting contours
TCGA-A2-A0YE: getting contours: non-roi: blood_vessel: adding contours
TCGA-A2-A0YE: getting contours: non-roi: angioinvasion: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: mostly_dcis: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: non-roi: other: NO OBJECTS!!
TCGA-A2-A0YE: getting contours: discarding backgrnd: discarded 3 contours
TCGA-A2-A0YE: getting contours: roi: roi: getting contours
TCGA-A2-A0YE: getting contours: roi: roi: adding contours
Let’s inspect the contours dataframe¶
The columns that really matter here are group, color, coords_x, and coords_y.
[9]:
contours_df.head()
[9]:
| group | color | ymin | ymax | xmin | xmax | has_holes | touches_edge-top | touches_edge-left | touches_edge-bottom | touches_edge-right | coords_x | coords_y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | roi | rgb(200,0,150) | 0.0 | 4593.0 | 0.0 | 4541.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 2835,2834,2833,2832,2831,2830,2829,2827,2826,2... | 0,1,1,2,2,3,3,5,5,6,6,8,8,9,9,10,10,12,12,13,1... |
| 1 | mostly_tumor | rgb(255,0,0) | 4269.0 | 4560.0 | 1639.0 | 2039.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1673,1672,1668,1667,1662,1661,1659,1658,1658,1... | 4269,4270,4270,4271,4271,4272,4272,4273,4274,4... |
| 2 | mostly_tumor | rgb(255,0,0) | 3764.0 | 4282.0 | 1607.0 | 2187.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1770,1769,1768,1767,1765,1764,1762,1761,1760,1... | 3764,3765,3765,3766,3766,3767,3767,3768,3768,3... |
| 3 | mostly_tumor | rgb(255,0,0) | 3712.0 | 4051.0 | 1201.0 | 1411.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1214,1213,1211,1210,1208,1207,1206,1205,1203,1... | 3712,3713,3713,3714,3714,3715,3715,3716,3716,3... |
| 4 | mostly_tumor | rgb(255,0,0) | 3356.0 | 3748.0 | 3108.0 | 3540.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3342,3341,3337,3336,3332,3331,3328,3327,3326,3... | 3356,3357,3357,3358,3358,3359,3359,3360,3360,3... |
3. Get annotation documents from contours¶
This method get_annotation_documents_from_contours() generates formatted annotation documents from contours that can be posted to the DSA server.
[10]:
print(get_annotation_documents_from_contours.__doc__)
Given dataframe of contours, get list of annotation documents.
This method parses a dataframe of contours to a list of dictionaries, each
of which represents and large_image style annotation. This is a wrapper
that extends the functionality of the method
get_single_annotation_document_from_contours(), whose docstring should
be referenced for implementation details and further explanation.
Parameters
-----------
contours_df : pandas DataFrame
WARNING - This is modified inside the function, so pass a copy.
This dataframe includes data on contours extracted from input mask
using get_contours_from_mask(). If you have contours using some other
method, just make sure the dataframe follows the same schema as the
output from get_contours_from_mask(). You may find a sample dataframe
in this repo at ./tests/test_files/annotations_and_masks/sample_contours_df.tsv
The following columns are relevant for this method.
group : str
annotation group (ground truth label).
color : str
annotation color if it were to be posted to DSA.
coords_x : str
vertex x coordinates comma-separated values
coords_y
vertex y coordinated comma-separated values
separate_docs_by_group : bool
if set to True, you get one or more annotation documents (dicts)
for each group (eg tumor) independently.
annots_per_doc : int
maximum number of annotation elements (polygons) per dict. The smaller
this number, the more numerous the annotation documents, but the more
seamless it is to post this data to the DSA server or to view using the
HistomicsTK interface since you will be loading smaller chunks of data
at a time.
annprops : dict
properties of annotation elements. Contains the following keys
F, X_OFFSET, Y_OFFSET, opacity, lineWidth. Refer to
get_single_annotation_document_from_contours() for details.
docnamePrefix : str
test to prepend to annotation document name
verbose : bool
Print progress to screen?
monitorPrefix : str
text to prepend to printed statements
Returns
--------
list of dicts
DSA-style annotation document.
As mentioned in the docs, this function wraps get_single_annotation_document_from_contours()
[11]:
print(get_single_annotation_document_from_contours.__doc__)
Given dataframe of contours, get annotation document.
This uses the large_image annotation schema to create an annotation
document that maybe posted to DSA for viewing using something like:
resp = gc.post("/annotation?itemId=" + slide_id, json=annotation_doc)
The annotation schema can be found at:
github.com/girder/large_image/blob/master/docs/annotations.md .
Parameters
-----------
contours_df_slice : pandas DataFrame
The following columns are of relevance and must be contained.
group : str
annotation group (ground truth label).
color : str
annotation color if it were to be posted to DSA.
coords_x : str
vertex x coordinates comma-separated values
coords_y
vertex y coordinated comma-separated values
docname : str
annotation document name
F : float
how much smaller is the mask where the contours come from is relative
to the slide scan magnification. For example, if the mask is at 10x
whereas the slide scan magnification is 20x, then F would be 2.0.
X_OFFSET : int
x offset to add to contours at BASE (SCAN) magnification
Y_OFFSET : int
y offset to add to contours at BASE (SCAN) magnification
opacity : float
opacity of annotation elements (in the range [0, 1])
lineWidth : float
width of boarders of annotation elements
verbose : bool
Print progress to screen?
monitorPrefix : str
text to prepend to printed statements
Returns
--------
dict
DSA-style annotation document ready to be post for viewing.
Let’s get a list of annotation documents (each is a dictionary). For the purpose of this tutorial, we separate the documents by group (i.e. each document is composed of polygons from the same style/group). You could decide to allow heterogeneous groups in the same annotation document by setting separate_docs_by_group to False. We place 10 polygons in each document for this demo for illustration purposes. Realistically you would want each document to contain several hundred depending on
their complexity. Placing too many polygons in each document can lead to performance issues when rendering in HistomicsUI.
Get annotation documents¶
[12]:
# get list of annotation documents
annprops = {
'X_OFFSET': X_OFFSET,
'Y_OFFSET': Y_OFFSET,
'opacity': 0.2,
'lineWidth': 4.0,
}
annotation_docs = get_annotation_documents_from_contours(
contours_df.copy(), separate_docs_by_group=True, annots_per_doc=10,
docnamePrefix='demo', annprops=annprops,
verbose=True, monitorPrefix=MASKNAME[:12] + ': annotation docs')
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 1 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 2 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 3 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 4 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 5 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 6 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 7 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 8 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 9 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 10 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 11 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 12 of 13
TCGA-A2-A0YE: annotation docs: mostly_lymphocytic_infiltrate: doc 1 of 1: contour 13 of 13
TCGA-A2-A0YE: annotation docs: exclude: doc 1 of 1: contour 1 of 1
TCGA-A2-A0YE: annotation docs: blood_vessel: doc 1 of 1: contour 1 of 3
TCGA-A2-A0YE: annotation docs: blood_vessel: doc 1 of 1: contour 2 of 3
TCGA-A2-A0YE: annotation docs: blood_vessel: doc 1 of 1: contour 3 of 3
TCGA-A2-A0YE: annotation docs: roi: doc 1 of 1: contour 1 of 1
TCGA-A2-A0YE: annotation docs: normal_acinus_or_duct: doc 1 of 1: contour 1 of 2
TCGA-A2-A0YE: annotation docs: normal_acinus_or_duct: doc 1 of 1: contour 2 of 2
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 1 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 2 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 3 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 4 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 5 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 6 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 7 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 8 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 9 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 1 of 2: contour 10 of 10
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 1 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 2 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 3 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 4 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 5 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 6 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 7 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 8 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 9 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 10 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 11 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 12 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 13 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 14 of 15
TCGA-A2-A0YE: annotation docs: mostly_tumor: doc 2 of 2: contour 15 of 15
Let’s examine one of the documents.¶
Limit display to the first two elements (polygons) and cap the vertices for clarity.
[13]:
ann_doc = annotation_docs[0].copy()
ann_doc['elements'] = ann_doc['elements'][:2]
for i in range(2):
ann_doc['elements'][i]['points'] = ann_doc['elements'][i]['points'][:5]
[14]:
ann_doc
[14]:
{'name': 'demo_mostly_lymphocytic_infiltrate-0',
'description': '',
'elements': [{'group': 'mostly_lymphocytic_infiltrate',
'type': 'polyline',
'lineColor': 'rgb(0,0,255)',
'lineWidth': 4.0,
'closed': True,
'points': [[61974.0, 37427.0, 0.0],
[61975.0, 37428.0, 0.0],
[61975.0, 37429.0, 0.0],
[61976.0, 37430.0, 0.0],
[61976.0, 37431.0, 0.0]],
'label': {'value': 'mostly_lymphocytic_infiltrate'},
'fillColor': 'rgba(0,0,255,0.2)'},
{'group': 'mostly_lymphocytic_infiltrate',
'type': 'polyline',
'lineColor': 'rgb(0,0,255)',
'lineWidth': 4.0,
'closed': True,
'points': [[60531.0, 37045.0, 0.0],
[60528.0, 37048.0, 0.0],
[60527.0, 37048.0, 0.0],
[60522.0, 37053.0, 0.0],
[60522.0, 37054.0, 0.0]],
'label': {'value': 'mostly_lymphocytic_infiltrate'},
'fillColor': 'rgba(0,0,255,0.2)'}]}
Post the annotation to the correct item/slide in DSA¶
[15]:
# deleting existing annotations in target slide (if any)
existing_annotations = gc.get('/annotation/item/' + SAMPLE_SLIDE_ID)
for ann in existing_annotations:
gc.delete('/annotation/%s' % ann['_id'])
# post the annotation documents you created
for annotation_doc in annotation_docs:
resp = gc.post(
'/annotation?itemId=' + SAMPLE_SLIDE_ID, json=annotation_doc)
Now you can go to HistomicsUI and confirm that the posted annotations make sense and correspond to tissue boundaries and expected labels.