Converting annotations to object segmentation mask images¶
Overview:
The DSA database stores annotations in an (x,y) coordinate list format. Some object localization algorithms like Faster-RCNN take coordinate formats whereas others (eg Mask R-CNN) require some form of object segmentation mask image whose pixel values encode not only class but instance information (so that individual objects of the same class can be distinguished).
This notebook demonstrates tools to convert annotations into contours or masks that can be used with algorithms like Mask-RCNN. There are two approaches for generating these data:
Generate contours or an object segmentation mask image from a region defined by user-specified coordinates.
Generate contours or object segmentation mask images from annotations contained within region-of-interest (ROI) annotations. This involves mapping annotations to these ROIs and creating one image per ROI.
The examples below extend approaches described in Amgad et al, 2019:
Mohamed Amgad, Habiba Elfandy, Hagar Hussein, …, Jonathan Beezley, Deepak R Chittajallu, David Manthey, David A Gutman, Lee A D Cooper, Structured crowdsourcing enables convolutional segmentation of histology images, Bioinformatics, , btz083, https://doi.org/10.1093/bioinformatics/btz083
A csv file like the one in histomicstk/annotations_and_masks/tests/test_files/sample_GTcodes.csv is needed to define what group each pixel value corresponds to in the mask image, to define the overlay order of various annotation groups, and which groups are considered to be ROIs. Note that the term “group” here comes from the annotation model where each group represents a class like “tumor” or “necrosis” and is associated with a an annotation style.
What is the difference between this and ``annotations_to_masks_handler``?
The difference between this and version 1, found at histomicstk.annotations_and_masks.annotations_to_masks_handler is that this (version 2) gets the contours first, including cropping to wanted ROI boundaries and other processing using shapely, and then parses these into masks. This enables us to differentiate various objects to use the data for object localization/classification/segmentation tasks. If you would like to get semantic segmentation masks instead, i.e. you do not care about
individual objects, you can use either version 1 or this handler using the semantic run mode. They re-use much of the same code-base, but some edge cases maybe better handled by version 1. For example, since this version uses shapely first to crop, some objects may be incorrectly parsed by shapely. Version 1, using PIL.ImageDraw may not have these problems.
Bottom line is: if you need semantic segmentation masks, it is probably safer to use version 1 (annotations to masks handler), whereas if you need object segmentation masks, this handler should be used in object run mode.
Where to look?
|_ histomicstk/
|_annotations_and_masks/
| |_annotation_and_mask_utils.py
| |_annotations_to_object_mask_handler.py
|_tests/
|_ test_annotation_and_mask_utils.py
|_ test_annotations_to_object_mask_handler.py
[1]:
import os
import copy
import girder_client
from pandas import DataFrame, read_csv
import tempfile
from imageio import imread
import matplotlib.pyplot as plt
%matplotlib inline
from histomicstk.annotations_and_masks.annotation_and_mask_utils import (
get_bboxes_from_slide_annotations,
scale_slide_annotations, get_scale_factor_and_appendStr)
from histomicstk.annotations_and_masks.annotations_to_object_mask_handler import (
annotations_to_contours_no_mask, contours_to_labeled_object_mask,
get_all_rois_from_slide_v2)
#Some nice default configuration for plots
plt.rcParams['figure.figsize'] = 7, 7
titlesize = 16
Connect girder client and set parameters¶
[2]:
CWD = os.getcwd()
APIURL = 'http://candygram.neurology.emory.edu:8080/api/v1/'
SAMPLE_SLIDE_ID = '5d586d57bd4404c6b1f28640'
GTCODE_PATH = os.path.join(CWD, '../../tests/test_files/sample_GTcodes.csv')
# connect to girder client
gc = girder_client.GirderClient(apiUrl=APIURL)
# gc.authenticate(interactive=True)
gc.authenticate(apiKey='kri19nTIGOkWH01TbzRqfohaaDWb6kPecRqGmemb')
# just a temp directory to save masks for now
BASE_SAVEPATH = tempfile.mkdtemp()
SAVEPATHS = {
'mask': os.path.join(BASE_SAVEPATH, 'masks'),
'rgb': os.path.join(BASE_SAVEPATH, 'rgbs'),
'contours': os.path.join(BASE_SAVEPATH, 'contours'),
'visualization': os.path.join(BASE_SAVEPATH, 'vis'),
}
for _, savepath in SAVEPATHS.items():
os.mkdir(savepath)
# What resolution do we want to get the images at?
# Microns-per-pixel / Magnification (either or)
MPP = 2.5 # <- this roughly translates to 4x magnification
MAG = None
Let’s inspect the ground truth codes file¶
This contains the ground truth codes and information dataframe. This is a dataframe that is indexed by the annotation group name and has the following columns:
group: group name of annotation (string), eg. “mostly_tumor”overlay_order: int, how early to place the annotation in the mask. Larger values means this annotation group is overlaid last and overwrites whatever overlaps it.GT_code: int, desired ground truth code (in the labeled mask) Pixels of this value belong to corresponding group (class)is_roi: Flag for whether this group marks ‘special’ annotations that encode the ROI boundaryis_background_class: Flag, whether this group is the default fill value inside the ROI. For example, you may decide that any pixel inside the ROI is considered stroma.
NOTE:
Zero pixels have special meaning and do not encode specific ground truth class. Instead, they simply mean ‘Outside ROI’ and should be ignored during model training or evaluation.
[3]:
# read GTCodes file
GTCodes_dict = read_csv(GTCODE_PATH)
GTCodes_dict.index = GTCodes_dict.loc[:, 'group']
GTCodes_dict = GTCodes_dict.to_dict(orient='index')
[4]:
GTCodes_dict.keys()
[4]:
dict_keys(['roi', 'evaluation_roi', 'mostly_tumor', 'mostly_stroma', 'mostly_lymphocytic_infiltrate', 'necrosis_or_debris', 'glandular_secretions', 'mostly_blood', 'exclude', 'metaplasia_NOS', 'mostly_fat', 'mostly_plasma_cells', 'other_immune_infiltrate', 'mostly_mucoid_material', 'normal_acinus_or_duct', 'lymphatics', 'undetermined', 'nerve', 'skin_adnexia', 'blood_vessel', 'angioinvasion', 'mostly_dcis', 'other'])
[5]:
GTCodes_dict['mostly_tumor']
[5]:
{'group': 'mostly_tumor',
'overlay_order': 1,
'GT_code': 1,
'is_roi': 0,
'is_background_class': 0,
'color': 'rgb(255,0,0)',
'comments': 'core class'}
Generate contours for user-defined region¶
Algorithms like Mask-RCNN consume coordinate data describing the boundaries of objects. The function annotations_to_contours_no_mask generates this contour data for user-specified region. These coordinate data in these contours is relative to the region frame instead of the whole-slide image frame.
[6]:
print(annotations_to_contours_no_mask.__doc__)
Process annotations to get RGB and contours without intermediate masks.
Parameters
----------
gc : object
girder client object to make requests, for example:
gc = girder_client.GirderClient(apiUrl = APIURL)
gc.authenticate(interactive=True)
slide_id : str
girder id for item (slide)
MPP : float or None
Microns-per-pixel -- best use this as it's more well-defined than
magnification which is more scanner or manufacturer specific.
MPP of 0.25 often roughly translates to 40x
MAG : float or None
If you prefer to use whatever magnification is reported in slide.
If neither MPP or MAG is provided, everything is retrieved without
scaling at base (scan) magnification.
mode : str
This specifies which part of the slide to get the mask from. Allowed
modes include the following
- wsi: get scaled up or down version of mask of whole slide
- min_bounding_box: get minimum box for all annotations in slide
- manual_bounds: use given ROI bounds provided by the 'bounds' param
- polygonal_bounds: use the idx_for_roi param to get coordinates
bounds : dict or None
if not None, has keys 'XMIN', 'XMAX', 'YMIN', 'YMAX' for slide
region coordinates (AT BASE MAGNIFICATION) to get labeled image
(mask) for. Use this with the 'manual_bounds' run mode.
idx_for_roi : int
index of ROI within the element_infos dataframe.
Use this with the 'polygonal_bounds' run mode.
slide_annotations : list or None
Give this parameter to avoid re-getting slide annotations. If you do
provide the annotations, though, make sure you have used
scale_slide_annotations() to scale them up or down by sf BEFOREHAND.
element_infos : pandas DataFrame.
The columns annidx and elementidx
encode the dict index of annotation document and element,
respectively, in the original slide_annotations list of dictionaries.
This can be obained by get_bboxes_from_slide_annotations() method.
Make sure you have used scale_slide_annotations().
linewidth : float
visualization line width
get_rgb: bool
get rgb image?
get_visualization : bool
get overlaid annotation bounds over RGB for visualization
text : bool
add text labels to visualization?
Returns
--------
dict
Results dict containing one or more of the following keys
- bounds: dict of bounds at scan magnification
- rgb: (mxnx3 np array) corresponding rgb image
- contours: dict
- visualization: (mxnx3 np array) visualization overlay
More input parameters¶
These optional parameters describe the desired magnification or resolution of the output, and whether to generate an RGB image for the region and a visualization image of the annotations.
[7]:
# common params for annotations_to_contours_no_mask()
annotations_to_contours_kwargs = {
'MPP': MPP, 'MAG': MAG,
'linewidth': 0.2,
'get_rgb': True,
'get_visualization': True,
'text': False,
}
1. manual_bounds mode¶
As shown in the example for generating semantic segmentation masks, this method can be run in four run modes: 1. wsi 2. min_bounding_box 3. manual_bounds 4. polygonal_bounds. Here we test the basic ‘manual_bounds’ mode where the boundaries of the region you want are provided at base/scan magnification.
[8]:
bounds = {
'XMIN': 58000, 'XMAX': 63000,
'YMIN': 35000, 'YMAX': 39000,
}
[9]:
# get specified region, let the method get and scale annotations
roi_out = annotations_to_contours_no_mask(
gc=gc, slide_id=SAMPLE_SLIDE_ID,
mode='manual_bounds', bounds=bounds,
**annotations_to_contours_kwargs)
/home/mtageld/Desktop/HistomicsTK/histomicstk/annotations_and_masks/annotation_and_mask_utils.py:668: RuntimeWarning: invalid value encountered in greater
iou = iou[:, iou[1, :] > iou_thresh].astype(int)
The result is an rgb image, contours and a visualization. Let’s take a look at these below.
[10]:
roi_out.keys()
[10]:
dict_keys(['rgb', 'contours', 'bounds', 'visualization'])
[11]:
roi_out['bounds']
[11]:
{'XMIN': 57994, 'XMAX': 62994, 'YMIN': 34999, 'YMAX': 38992}
[12]:
for imstr in ['rgb', 'visualization']:
plt.imshow(roi_out[imstr])
plt.title(imstr)
plt.show()
[13]:
DataFrame(roi_out['contours']).head()
[13]:
| annidx | annotation_girder_id | elementidx | element_girder_id | type | group | label | color | xmin | xmax | ymin | ymax | bbox_area | coords_x | coords_y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 5d586d58bd4404c6b1f28642 | 0 | 5a943eb992ca9a0016fae97f | rectangle | roi | roi | rgb(163, 19, 186) | 121 | 501 | 0 | 310 | 117800 | 491,143,121,292,501,501,491 | 0,0,13,310,189,15,0 |
| 1 | 1 | 5d586d58bd4404c6b1f28644 | 0 | 5a943eb992ca9a0016fae981 | polyline | blood_vessel | blood_vessel | rgb(128,0,128) | 341 | 360 | 241 | 259 | 342 | 353,352,351,350,350,349,348,347,346,345,344,34... | 241,241,241,241,242,242,242,242,242,242,242,24... |
| 2 | 1 | 5d586d58bd4404c6b1f28644 | 1 | 5a943eb992ca9a0016fae982 | polyline | blood_vessel | blood_vessel | rgb(128,0,128) | 244 | 258 | 161 | 181 | 280 | 244,244,244,244,244,244,245,245,245,245,245,24... | 161,162,163,165,166,167,167,168,169,170,171,17... |
| 3 | 2 | 5d586d58bd4404c6b1f2864c | 0 | 5a943eba92ca9a0016fae990 | polyline | mostly_lymphocytic_infiltrate | mostly_lymphocytic_infiltrate | rgb(0,0,255) | 388 | 501 | 71 | 236 | 18645 | 501,501,500,500,499,498,498,497,496,495,495,49... | 171,75,75,74,74,74,73,73,72,72,71,71,71,71,72,... |
| 4 | 2 | 5d586d58bd4404c6b1f2864c | 1 | 5a943eba92ca9a0016fae994 | polyline | mostly_lymphocytic_infiltrate | mostly_lymphocytic_infiltrate | rgb(0,0,255) | 359 | 434 | 0 | 21 | 1575 | 434,359,359,359,360,360,360,361,361,362,362,36... | 0,0,1,2,2,3,4,5,6,6,7,7,7,7,7,8,8,8,9,9,11,12,... |
Note that if the above function call is made repeatedly for the same slide (e.g. to iterate over multiple regions), multiple get requests would be created to retrieve annotations from the server. To improve efficiency when handling multiple regions in the same slide, we could manually get annotations and scale them down/up to desired resolution, and pass them to annotations_to_contours_no_mask().
[14]:
# get annotations for slide
slide_annotations = gc.get('/annotation/item/' + SAMPLE_SLIDE_ID)
# scale up/down annotations by a factor
sf, _ = get_scale_factor_and_appendStr(
gc=gc, slide_id=SAMPLE_SLIDE_ID, MPP=MPP, MAG=MAG)
slide_annotations = scale_slide_annotations(slide_annotations, sf=sf)
# get bounding box information for all annotations
element_infos = get_bboxes_from_slide_annotations(slide_annotations)
[15]:
# get specified region -- manually providing scaled annotations
roi_out = annotations_to_contours_no_mask(
gc=gc, slide_id=SAMPLE_SLIDE_ID,
mode='manual_bounds', bounds=bounds,
slide_annotations=slide_annotations, element_infos=element_infos,
**annotations_to_contours_kwargs)
/home/mtageld/Desktop/HistomicsTK/histomicstk/annotations_and_masks/annotation_and_mask_utils.py:668: RuntimeWarning: invalid value encountered in greater
iou = iou[:, iou[1, :] > iou_thresh].astype(int)
[16]:
roi_out['bounds']
[16]:
{'XMIN': 57994, 'XMAX': 62994, 'YMIN': 34999, 'YMAX': 38992}
2. min_bounding_box mode¶
In min_bounding_box mode everything in the slide is handled using the smallest rectangular bounding box.
[17]:
# get ROI bounding everything
minbbox_out = annotations_to_contours_no_mask(
gc=gc, slide_id=SAMPLE_SLIDE_ID,
mode='min_bounding_box', **annotations_to_contours_kwargs)
[18]:
minbbox_out['bounds']
[18]:
{'XMIN': 56726, 'YMIN': 33483, 'XMAX': 63732, 'YMAX': 39890}
[19]:
plt.imshow(minbbox_out['visualization'])
[19]:
<matplotlib.image.AxesImage at 0x7f1825c3f8d0>
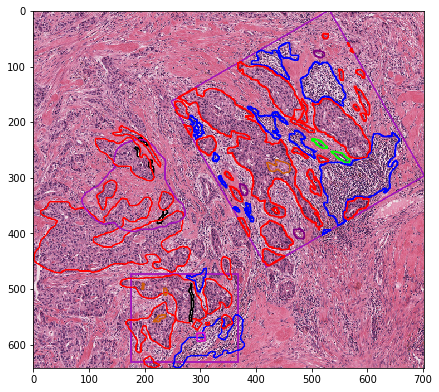
3. wsi mode¶
wsi mode creates a scaled version of the entire whole-slide image and all annotations contained within.
NOTE:
This does not rely on tiles and processes the image at whatever magnification you want. You can suppress the RGB or visualization outputs and to just fetch the contours or object segmentation mask (see below), providing a bigger magnification range before encountering memory problems.
[20]:
# get entire wsi region
get_kwargs = copy.deepcopy(annotations_to_contours_kwargs)
get_kwargs['MPP'] = 5.0 # otherwise it's too large!
wsi_out = annotations_to_contours_no_mask(
gc=gc, slide_id=SAMPLE_SLIDE_ID,
mode='wsi', **get_kwargs)
[21]:
wsi_out['bounds']
[21]:
{'XMIN': 0, 'XMAX': 131516, 'YMIN': 0, 'YMAX': 80439}
[22]:
plt.imshow(wsi_out['visualization'])
plt.show()
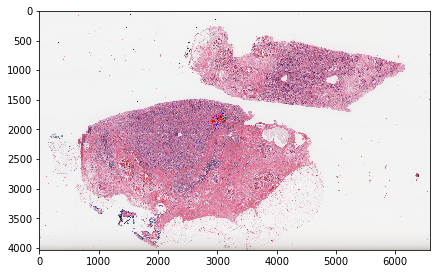
[23]:
plt.imshow(wsi_out['visualization'][1500:2000, 2800:3300])
plt.show()
Parse manually-drawn ROIs into separate labeled object masks¶
This function utilizes the polygonal_bounds mode of the get_image_and_mask_from_slide() method to generate a set of outputs for each ROI annotation.
In the example above we focused on generating contour data to represent objects. Below we focus on generating object segmentation mask images.
[24]:
print(get_all_rois_from_slide_v2.__doc__)
Get all ROIs for a slide without an intermediate mask form.
This mainly relies on contours_to_labeled_object_mask(), which should
be referred to for extra documentation.
This can be run in either the "object" mode, whereby the saved masks
are a three-channel png where first channel encodes class label (i.e.
same as semantic segmentation) and the product of the values in the
second and third channel encodes the object ID. Otherwise, the user
may decide to run in the "semantic" mode and the resultant mask would
consist of only one channel (semantic segmentation with no object
differentiation).
The difference between this and version 1, found at
histomicstk.annotations_and_masks.annotations_to_masks_handler.
get_all_rois_from_slide()
is that this (version 2) gets the contours first, including cropping
to wanted ROI boundaries and other processing using shapely, and THEN
parses these into masks. This enables us to differentiate various objects
to use the data for object localization or classification or segmentation
tasks. If you would like to get semantic segmentation masks, i.e. you do
not really care about individual objects, you can use either version 1
or this method. They re-use much of the same code-base, but some edge
cases maybe better handled by version 1. For example, since
this version uses shapely first to crop, some objects may be incorrectly
parsed by shapely. Version 1, using PIL.ImageDraw may not have these
problems.
Bottom line is: if you need semantic segmentation masks, it is probably
safer to use version 1, whereas if you need object segmentation masks,
this method should be used.
Parameters
----------
gc : object
girder client object to make requests, for example:
gc = girder_client.GirderClient(apiUrl = APIURL)
gc.authenticate(interactive=True)
slide_id : str
girder id for item (slide)
GTCodes_dict : dict
the ground truth codes and information dict.
This is a dict that is indexed by the annotation group name and
each entry is in turn a dict with the following keys:
- group: group name of annotation (string), eg. mostly_tumor
- overlay_order: int, how early to place the annotation in the
mask. Larger values means this annotation group is overlaid
last and overwrites whatever overlaps it.
- GT_code: int, desired ground truth code (in the mask)
Pixels of this value belong to corresponding group (class)
- is_roi: Flag for whether this group encodes an ROI
- is_background_class: Flag, whether this group is the default
fill value inside the ROI. For example, you may decide that
any pixel inside the ROI is considered stroma.
save_directories : dict
paths to directories to save data. Each entry is a string, and the
following keys are allowed
- ROI: path to save masks (labeled images)
- rgb: path to save rgb images
- contours: path to save annotation contours
- visualization: path to save rgb visualization overlays
mode : str
run mode for getting masks. Must me in
- object: get 3-channel mask where first channel encodes label
(tumor, stroma, etc) while product of second and third
channel encodes the object ID (i.e. individual contours)
This is useful for object localization and segmentation tasks.
- semantic: get a 1-channel mask corresponding to the first channel
of the object mode.
get_mask : bool
While the main purpose of this method IS to get object segmentation
masks, it is conceivable that some users might just want to get
the RGB and contours. Default is True.
annotations_to_contours_kwargs : dict
kwargs to pass to annotations_to_contours_no_mask()
default values are assigned if specific parameters are not given.
slide_name : str or None
If not given, its inferred using a server request using girder client.
verbose : bool
Print progress to screen?
monitorprefix : str
text to prepend to printed statements
callback : function
a callback function to run on the roi dictionary output. This is
internal, but if you really want to use this, make sure the callback
can accept the following keys and that you do NOT assign them yourself
gc, slide_id, slide_name, MPP, MAG, verbose, monitorprefix
Also, this callback MUST *ONLY* return thr roi dictionary, whether
or not it is modified inside it. If it is modified inside the callback
then the modified version is the one that will be saved to disk.
callback_kwargs : dict
kwargs to pass to callback, not including the mandatory kwargs
that will be passed internally (mentioned earlier here).
Returns
--------
list of dicts
each entry contains the following keys
mask - path to saved mask
rgb - path to saved rgb image
contours - path to saved annotation contours
visualization - path to saved rgb visualization overlay
The above method mainly relies on contours_to_labeled_object_mask(), described below.
[25]:
print(contours_to_labeled_object_mask.__doc__)
Process contours to get and object segmentation labeled mask.
Parameters
----------
contours : DataFrame
contours corresponding to annotation elemeents from the slide.
All coordinates are relative to the mask that you want to output.
The following columns are expected.
- group: str, annotation group (ground truth label).
- ymin: int, minimum y coordinate
- ymax: int, maximum y coordinate
- xmin: int, minimum x coordinate
- xmax: int, maximum x coordinate
- coords_x: str, vertex x coordinates comma-separated values
- coords_y: str, vertex y coordinated comma-separated values
gtcodes : DataFrame
the ground truth codes and information dataframe.
This is a dataframe that is indexed by the annotation group name
and has the following columns.
- group: str, group name of annotation, eg. mostly_tumor.
- GT_code: int, desired ground truth code (in the mask).
Pixels of this value belong to corresponding group (class).
- color: str, rgb format. eg. rgb(255,0,0).
mode : str
run mode for getting masks. Must be in
- object: get 3-channel mask where first channel encodes label
(tumor, stroma, etc) while product of second and third
channel encodes the object ID (i.e. individual contours)
This is useful for object localization and segmentation tasks.
- semantic: get a 1-channel mask corresponding to the first channel
of the object mode.
verbose : bool
print to screen?
monitorprefix : str
prefix to add to printed statements
Returns
-------
np.array
If mode is "object", this returns an (m, n, 3) np array
that can be saved as a png
First channel: encodes label (can be used for semantic segmentation)
Second & third channels: multiplication of second and third channel
gives the object id (255 choose 2 = 32,385 max unique objects).
This allows us to save into a convenient 3-channel png object labels
and segmentation masks, which is more compact than traditional
mask-rcnn save formats like having one channel per object and a
separate csv file for object labels. This is also more convenient
than simply saving things into pickled np array objects, and allows
compatibility with data loaders that expect an image or mask.
If mode is "semantic" only the labels (corresponding to first
channel of the object mode) is output.
[26]:
get_all_rois_kwargs = {
'gc': gc,
'slide_id': SAMPLE_SLIDE_ID,
'GTCodes_dict': GTCodes_dict,
'save_directories': SAVEPATHS,
'annotations_to_contours_kwargs': annotations_to_contours_kwargs,
'slide_name': 'TCGA-A2-A0YE',
'mode': 'object',
'verbose': True,
'monitorprefix': 'test',
}
savenames = get_all_rois_from_slide_v2(**get_all_rois_kwargs)
test: roi 1 of 3: Overlay level -1: Element 1 of 50: roi
test: roi 1 of 3: Overlay level 1: Element 2 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 3 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 4 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 5 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 6 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 7 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 8 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 9 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 10 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 11 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 12 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 13 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 14 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 15 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 16 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 17 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 18 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 19 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 20 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 21 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 22 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 23 of 50: normal_acinus_or_duct
test: roi 1 of 3: Overlay level 1: Element 24 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 25 of 50: normal_acinus_or_duct
test: roi 1 of 3: Overlay level 1: Element 26 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 27 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 28 of 50: blood_vessel
test: roi 1 of 3: Overlay level 1: Element 29 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 30 of 50: blood_vessel
test: roi 1 of 3: Overlay level 1: Element 31 of 50: blood_vessel
test: roi 1 of 3: Overlay level 1: Element 32 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 33 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 34 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 35 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 36 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 37 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 38 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 39 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 40 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 41 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 42 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 43 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 44 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 45 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 1: Element 46 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 47 of 50: mostly_lymphocytic_infiltrate
test: roi 1 of 3: Overlay level 1: Element 48 of 50: mostly_tumor
test: roi 1 of 3: Overlay level 2: Element 49 of 50: mostly_stroma
test: roi 1 of 3: Overlay level 3: Element 50 of 50: exclude
test: roi 1 of 3: Saving /tmp/tmpvdwsnssq/masks/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.png
test: roi 1 of 3: Saving /tmp/tmpvdwsnssq/rgbs/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.png
test: roi 1 of 3: Saving /tmp/tmpvdwsnssq/vis/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.png
test: roi 1 of 3: Saving /tmp/tmpvdwsnssq/contours/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.csv
test: roi 2 of 3: Overlay level -2: Element 1 of 16: roi
test: roi 2 of 3: Overlay level 1: Element 2 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 3 of 16: mostly_lymphocytic_infiltrate
test: roi 2 of 3: Overlay level 1: Element 4 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 5 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 6 of 16: mostly_lymphocytic_infiltrate
test: roi 2 of 3: Overlay level 1: Element 7 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 8 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 9 of 16: blood_vessel
test: roi 2 of 3: Overlay level 1: Element 10 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 11 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 1: Element 12 of 16: mostly_tumor
test: roi 2 of 3: Overlay level 2: Element 13 of 16: mostly_stroma
test: roi 2 of 3: Overlay level 2: Element 14 of 16: mostly_stroma
test: roi 2 of 3: Overlay level 2: Element 15 of 16: mostly_stroma
test: roi 2 of 3: Overlay level 3: Element 16 of 16: exclude
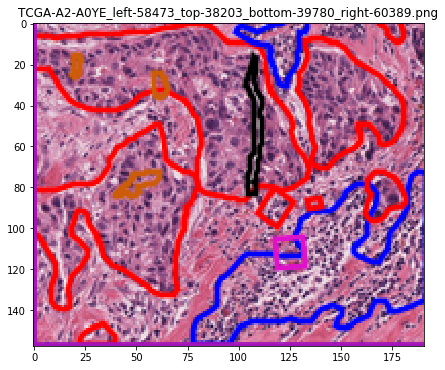
test: roi 2 of 3: Saving /tmp/tmpvdwsnssq/masks/TCGA-A2-A0YE_left-58473_top-38203_bottom-39780_right-60389.png
test: roi 2 of 3: Saving /tmp/tmpvdwsnssq/rgbs/TCGA-A2-A0YE_left-58473_top-38203_bottom-39780_right-60389.png
test: roi 2 of 3: Saving /tmp/tmpvdwsnssq/vis/TCGA-A2-A0YE_left-58473_top-38203_bottom-39780_right-60389.png
test: roi 2 of 3: Saving /tmp/tmpvdwsnssq/contours/TCGA-A2-A0YE_left-58473_top-38203_bottom-39780_right-60389.csv
test: roi 3 of 3: Overlay level -3: Element 1 of 11: roi
test: roi 3 of 3: Overlay level 1: Element 2 of 11: mostly_tumor
test: roi 3 of 3: Overlay level 1: Element 3 of 11: mostly_tumor
test: roi 3 of 3: Overlay level 1: Element 4 of 11: mostly_tumor
test: roi 3 of 3: Overlay level 1: Element 5 of 11: mostly_tumor
test: roi 3 of 3: Overlay level 1: Element 6 of 11: mostly_tumor
test: roi 3 of 3: Overlay level 1: Element 7 of 11: mostly_tumor
test: roi 3 of 3: Overlay level 3: Element 8 of 11: exclude
test: roi 3 of 3: Overlay level 3: Element 9 of 11: exclude
test: roi 3 of 3: Overlay level 3: Element 10 of 11: exclude
test: roi 3 of 3: Overlay level 3: Element 11 of 11: exclude
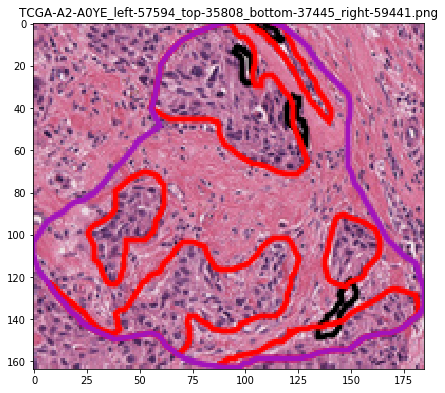
test: roi 3 of 3: Saving /tmp/tmpvdwsnssq/masks/TCGA-A2-A0YE_left-57594_top-35808_bottom-37445_right-59441.png
test: roi 3 of 3: Saving /tmp/tmpvdwsnssq/rgbs/TCGA-A2-A0YE_left-57594_top-35808_bottom-37445_right-59441.png
test: roi 3 of 3: Saving /tmp/tmpvdwsnssq/vis/TCGA-A2-A0YE_left-57594_top-35808_bottom-37445_right-59441.png
test: roi 3 of 3: Saving /tmp/tmpvdwsnssq/contours/TCGA-A2-A0YE_left-57594_top-35808_bottom-37445_right-59441.csv
[27]:
savenames[0]
[27]:
{'mask': '/tmp/tmpvdwsnssq/masks/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.png',
'rgb': '/tmp/tmpvdwsnssq/rgbs/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.png',
'visualization': '/tmp/tmpvdwsnssq/vis/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.png',
'contours': '/tmp/tmpvdwsnssq/contours/TCGA-A2-A0YE_left-59191_top-33483_bottom-38083_right-63732.csv'}
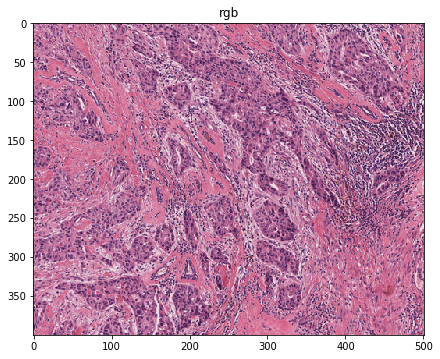
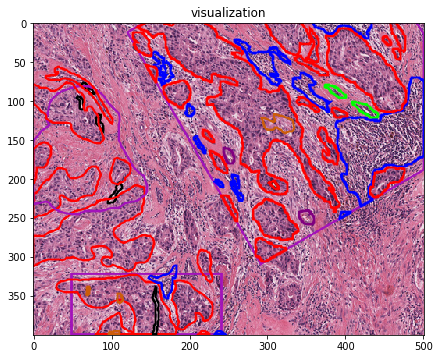
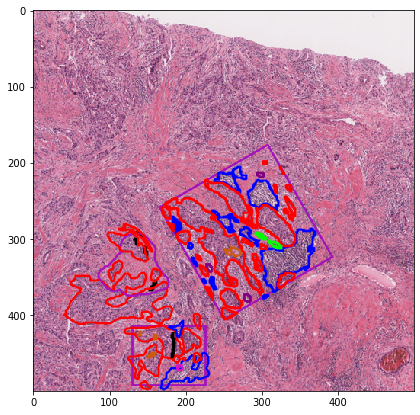
Let’s visualize the contours¶
[28]:
# visualization of contours over RGBs
for savename in savenames:
vis = imread(savename['visualization'])
plt.imshow(vis)
plt.title(os.path.basename(savename['visualization']))
plt.show()
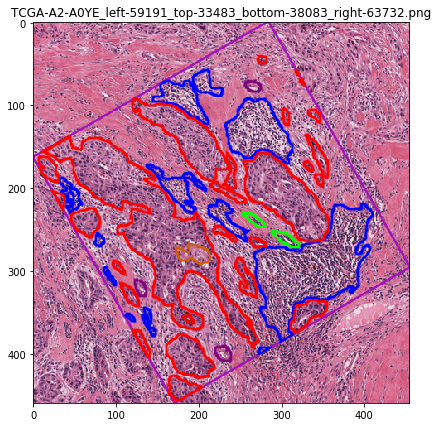
Let’s visualize the object mask¶
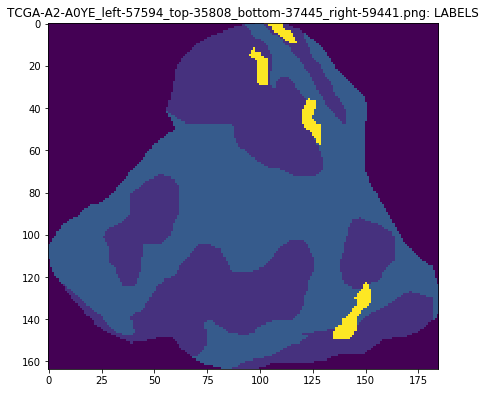
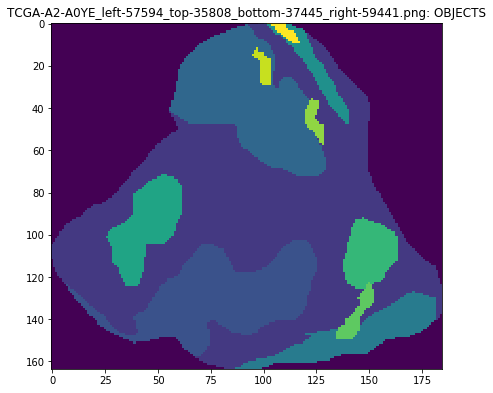
An object segmentation mask image uses the multiple channels to encode both class and instance information. In this mask format we multiply the second and third channel values to calculate a unique id for each object.
[29]:
mask = imread(savename['mask'])
maskname = os.path.basename(savename['mask'])
plt.imshow(mask[..., 0])
plt.title(maskname + ': LABELS')
plt.show()
plt.imshow(mask[..., 1] * mask[..., 2])
plt.title(maskname + ': OBJECTS')
plt.show()