The Story
The primary determinant in machine learning algorithm performance is the availability of abundant labeled data for training. Collecting data for applications like tissue region segmentation and cell classification is challenging given the limited availability of experts and tools needed to collect and review annotations.
Our web-based platform enables users to collaborate all over the world, and has been used to generate over 120,000 human markups of histopathology in multiple annotation studies.
Our API and user roles allow us to create collaborative annotation study teams that engage users with different expertise levels, ranging from pathologists to medical students, to collaboratively generate large and extensively reviewed annotation datasets. In a study with over 25 participants from over 5 countries, we generated over 25,000 annotated tissue regions to generate the richest public dataset of annotated breast-cancer tissues to date. The ability to programmatically monitor and manage these studies through the API is key to their success.
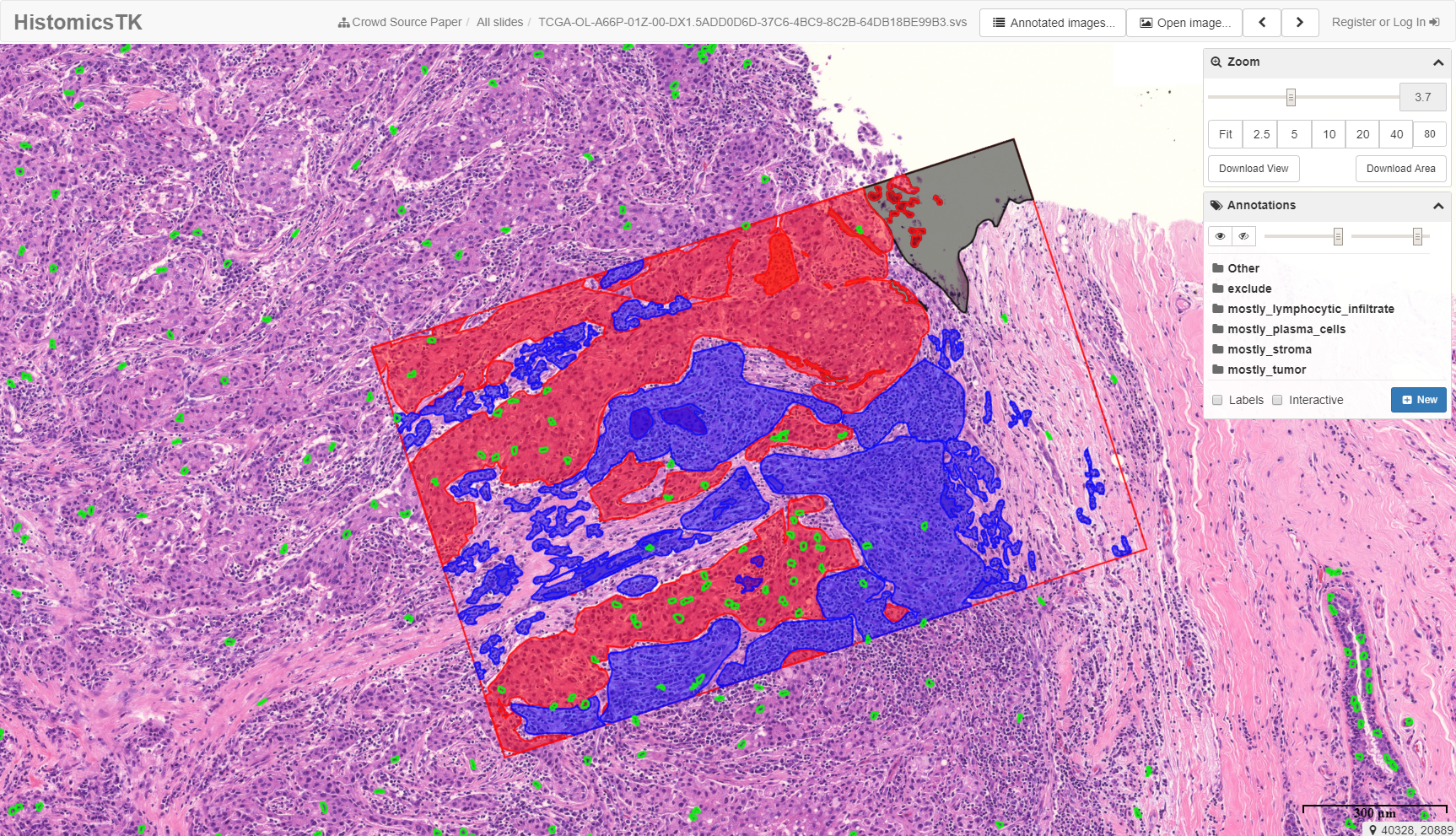
Collecting data on this scale allows us to generate highly accurate machine-learning models for tasks like tissue segmentation and cell classification and detection, and to understand the limits of concordance among human experts. The data from the collaborative annotation study is available on a demo instance of the Digital Slide Archive. The link goes to one of the TCGA slides that was used in the study. If you click the “eye” image icon in the Annotations panel on the right side of the screen, you’ll see the results of a collaborative annotation.
References
Chandradevan, Ramraj, Ahmed A. Aljudi, Bradley R. Drumheller, Nilakshan Kunananthaseelan, Mohamed Amgad, David A. Gutman, Lee A. D. Cooper, and David L. Jaye. “Machine-Based Detection and Classification for Bone Marrow Aspirate Differential Counts: Initial Development Focusing on Nonneoplastic Cells.” Laboratory Investigation 100, no. 1 (September 30, 2019): 98–109. https://doi.org/10.1038/s41374-019-0325-7
PMID: 31570774
Amgad, Mohamed, Habiba Elfandy, Hagar Hussein, Lamees A Atteya, Mai A T Elsebaie, Lamia S Abo Elnasr, Rokia A Sakr, et al. “Structured Crowdsourcing Enables Convolutional Segmentation of Histology Images.” Edited by Robert Murphy. Bioinformatics 35, no. 18 (February 6, 2019): 3461–67. https://doi.org/10.1093/bioinformatics/btz083
PMID: 30726865
Mobadersany, Pooya, Safoora Yousefi, Mohamed Amgad, David A. Gutman, Jill S. Barnholtz-Sloan, José E. Velázquez Vega, Daniel J. Brat, and Lee A. D. Cooper. “Predicting Cancer Outcomes from Histology and Genomics Using Convolutional Networks.” Proceedings of the National Academy of Sciences 115, no. 13 (March 12, 2018): E2970–79. https://doi.org/10.1073/pnas.1717139115
PMID: 29531073