What is DSA?
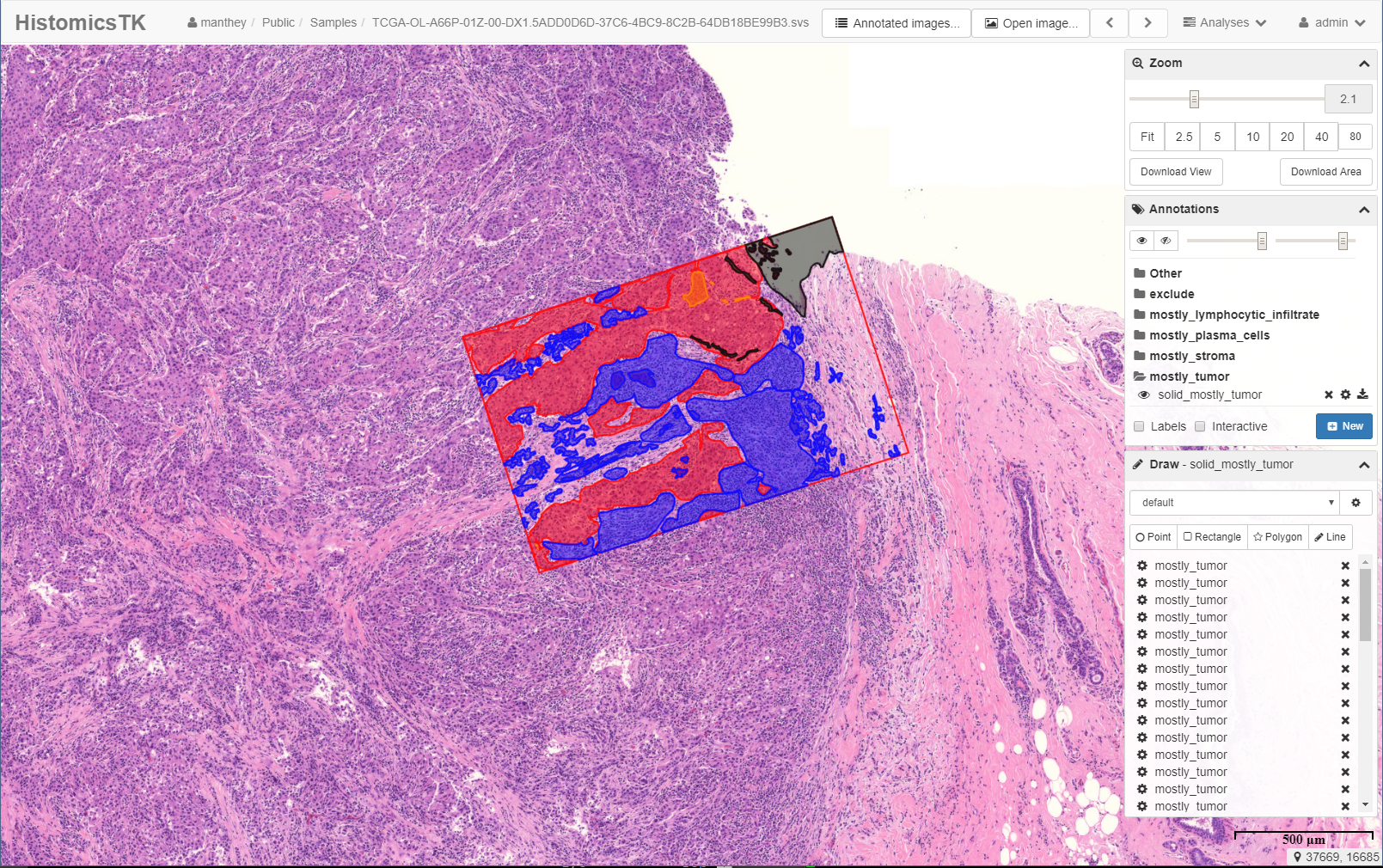
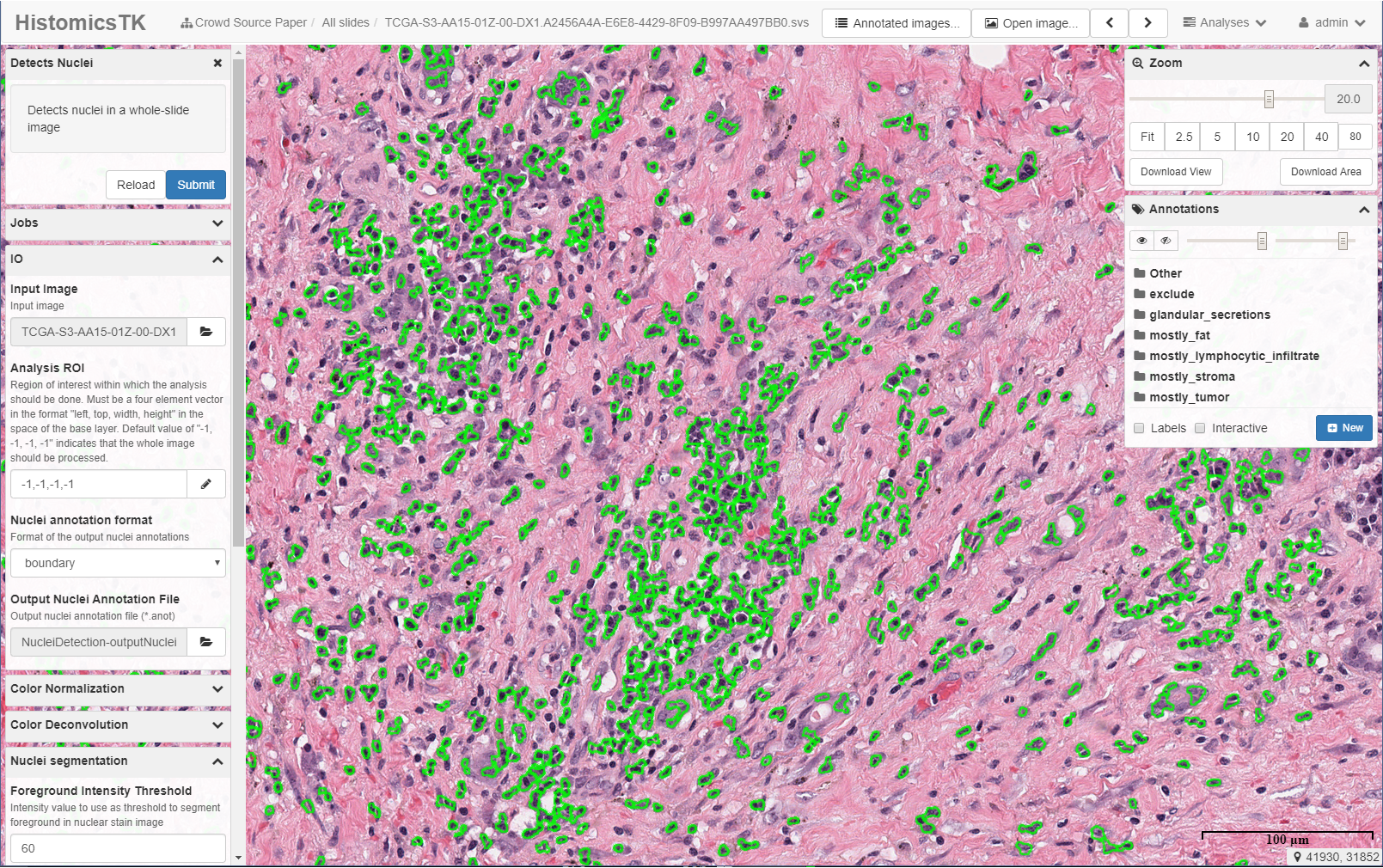
The Digital Slide Archive (DSA) is a platform that provides the ability to store, manage, visualize and annotate large imaging data sets. The DSA consists of an analysis toolkit (HistomicsTK), an interface to visualize slides and manage annotations (HistomicsUI), a database layer (using Mongo), and a web-server that provides a rich API and data management tools (using Girder). This system can:
- Organize images from a variety of assetstores, such as local files systems and S3.
- Provide user access controls.
- Image annotation and review.
- Run algorithms on all or parts of images.